Risk Model for Early Detection of Parkinson's Disease
Project in Progress
Preliminary Code and Graphs Provided
Developing Predictive Models
# Import libraries
import numpy as np
import pandas as pd
from time import time
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
from sklearn.naive_bayes import GaussianNB
from sklearn import svm
from sklearn.linear_model import SGDClassifier
from sklearn.ensemble import GradientBoostingClassifier
import matplotlib.pyplot as plt
from pandas.plotting import scatter_matrix
# Training and Testing Functions
def train_classifier(clf, X_train, y_train):
''' Fits a classifier to the training data. '''
start = time()
clf.fit(X_train, y_train)
end = time()
print("Trained model in {:.4f} seconds".format(end - start))
def predict_labels(clf, features, target):
''' Makes predictions using a fit classifier based on F1 score. '''
start = time()
y_pred = clf.predict(features)
end = time()
print("Made predictions in {:.4f} seconds.".format(end - start))
return f1_score(target.values, y_pred, pos_label=1)
def train_predict(clf, X_train, y_train, X_test, y_test):
''' Train and predict using a classifier based on F1 score. '''
print("Training a {} using a training set size of {}. . .".format(clf.__class__.__name__, len(X_train)))
train_classifier(clf, X_train, y_train)
print("F1 score for training set: {:.4f}.".format(predict_labels(clf, X_train, y_train)))
print("F1 score for test set: {:.4f}.".format(predict_labels(clf, X_test, y_test)))
# Tuning / Optimization Functions
def performance_metric(y_true, y_predict):
error = f1_score(y_true, y_predict, pos_label=1)
return error
def fit_model(X, y):
classifier = svm.SVC()
parameters = {'kernel': ['poly', 'rbf', 'sigmoid'], 'degree': [1, 2, 3], 'C': [0.1, 1, 10]}
f1_scorer = make_scorer(performance_metric, greater_is_better=True)
clf = GridSearchCV(classifier, param_grid=parameters, scoring=f1_scorer)
clf.fit(X, y)
return clf
# Read parkinson data
parkinson_data = pd.read_csv("parkinsons.csv")
print("Parkinson data read successfully!")
# Data Exploration
n_patients = parkinson_data.shape[0]
n_features = parkinson_data.shape[1] - 1
n_parkinsons = parkinson_data[parkinson_data['status'] == 1].shape[0]
n_healthy = parkinson_data[parkinson_data['status'] == 0].shape[0]
print("Total number of patients: {}".format(n_patients))
print("Number of features: {}".format(n_features))
print("Number of patients with Parkinson's: {}".format(n_parkinsons))
print("Number of patients without Parkinson's: {}".format(n_healthy))
# Preparing the Data
feature_cols = list(parkinson_data.columns[1:16]) + list(parkinson_data.columns[18:])
target_col = parkinson_data.columns[17]
X_all = parkinson_data[feature_cols]
y_all = parkinson_data[target_col]
print("\nFeature columns:\n{}".format(feature_cols))
print("\nTarget column: {}".format(target_col))
print("\nFeature values:")
print(X_all.head())
num_all = parkinson_data.shape[0]
num_train = 150
num_test = num_all - num_train
X_train, X_test, y_train, y_test = train_test_split(X_all, y_all, test_size=num_test, random_state=5)
print("Shuffling of data into test and training sets complete!")
print("Training set: {} samples".format(X_train.shape[0]))
print("Test set: {} samples".format(X_test.shape[0]))
X_train_50 = X_train[:50]
y_train_50 = y_train[:50]
X_train_100 = X_train[:100]
y_train_100 = y_train[:100]
X_train_150 = X_train[:150]
y_train_150 = y_train[:150]
# Training the data
clf = GaussianNB()
clf2 = svm.SVC()
clf3 = SGDClassifier(loss="hinge")
clf4 = GradientBoostingClassifier(n_estimators=100, learning_rate=1.0, max_depth=1, random_state=0)
print("50 samples:")
train_predict(clf, X_train_50, y_train_50, X_test, y_test)
train_predict(clf2, X_train_50, y_train_50, X_test, y_test)
train_predict(clf3, X_train_50, y_train_50, X_test, y_test)
train_predict(clf4, X_train_50, y_train_50, X_test, y_test)
print("\n100 samples:")
train_predict(clf, X_train_100, y_train_100, X_test, y_test)
train_predict(clf2, X_train_100, y_train_100, X_test, y_test)
train_predict(clf3, X_train_100, y_train_100, X_test, y_test)
train_predict(clf4, X_train_100, y_train_100, X_test, y_test)
print("\n150 samples:")
train_predict(clf, X_train_150, y_train_150, X_test, y_test)
train_predict(clf2, X_train_150, y_train_150, X_test, y_test)
train_predict(clf3, X_train_150, y_train_150, X_test, y_test)
train_predict(clf4, X_train_150, y_train_150, X_test, y_test)
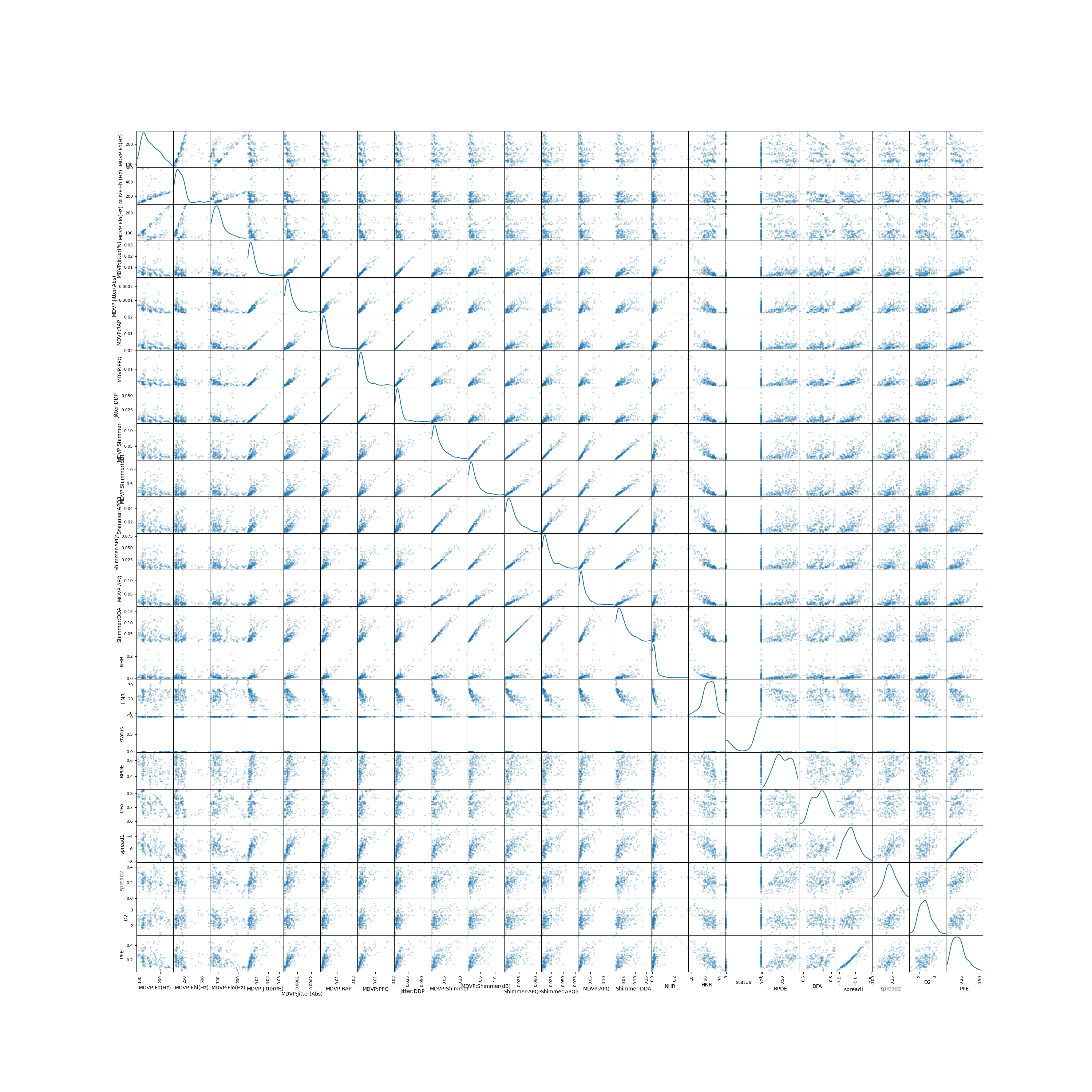
# Data Visualization
scatter_matrix(parkinson_data, alpha=0.3, figsize=(30, 30), diagonal='kde')
plt.savefig("scatter.png")
# Tuning the model
print("Tuning the model. This may take a while.....")
clf2 = fit_model(X_train, y_train)
print("Successfully fit a model!")
print("The best parameters were:")
print(clf2.best_params_)
start = time()
print("Tuned model has a training F1 score of {:.4f}.".format(predict_labels(clf2, X_train, y_train)))
print("Tuned model has a testing F1 score of {:.4f}.".format(predict_labels(clf2, X_test, y_test)))
end = time()
print("Tuned model in {:.4f} seconds.".format(end - start))